Exercise 8b - Enzyme Design
Exercises | | Links:
We will look at some examples of catalysis and try our luck in designing some enzymes ourselves!
Catalytic antibodies
An antibody that specifically binds the chemical compound B also catalyzes the degradation of compounds 3-5 as indicated below.

Task 1: Explain how the antibody can catalyze these reactions.
The antibody has so-called “broad substrate specificity”, ie. it can degrade very different substances. To investigate how this can be done, an attempt was made to solve the structure of the antibody in complex with substance 1, but this proved unsuccessful. However, crystals of the complex between the antibody and compound 2 (PDB structure 5XQW) were obtained. The structure is given below, where the heavy chain of the antibody is shown in green, the light chain in turquoise and compound 2 in yellow (for the carbon atoms).
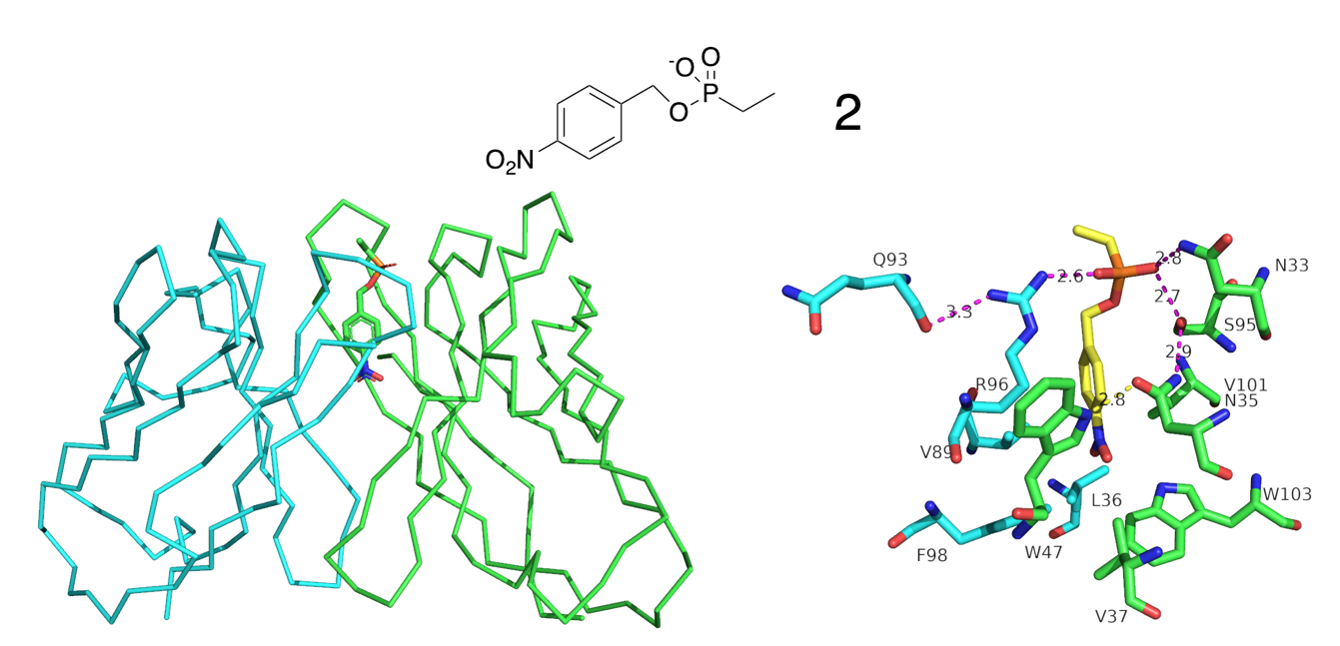
Task 2: Based on the structure, explain why the antibody has broad specificity.
Biological plastic degradation
The last 50-70 years have witnessed a colossal accumulation of synthetically produced plastic as waste in nature. Mostly this waste is buried in landfills or incinerated. However, a large part is dispersed in Nature, where it is slowly physically broken down into smaller particles. Very little enzyme-driven degradation of plastic takes place. The structures of the most common classes of plastics are shown below.

Task 3: From a chemical perspective, why is it a challenge for enzymes to degrade these substances? Which compound groups are expected to be hardest and easiest to degrade and why? What does this have to do with drug design?
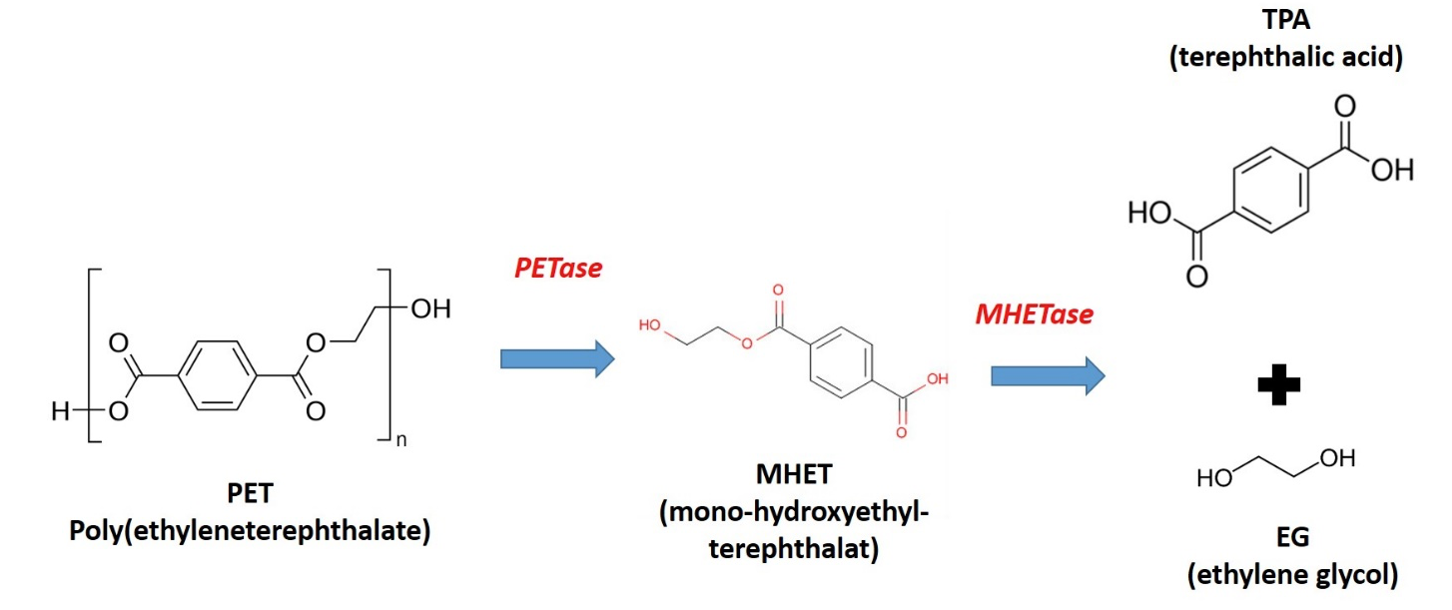
Nevertheless, various enzymes have been identified that (albeit slowly) degrade different types of plastic. Some of these are produced by microorganisms that grow in a landfill for primarily PET plastic. One enzyme (PETase) degrades PET to MHET, while another enzyme (a MHETase) degrades MHET to PTA and EG (see reaction scheme below).
The structure of MHETase in the presence of the compound benzoate has been solved (see below).
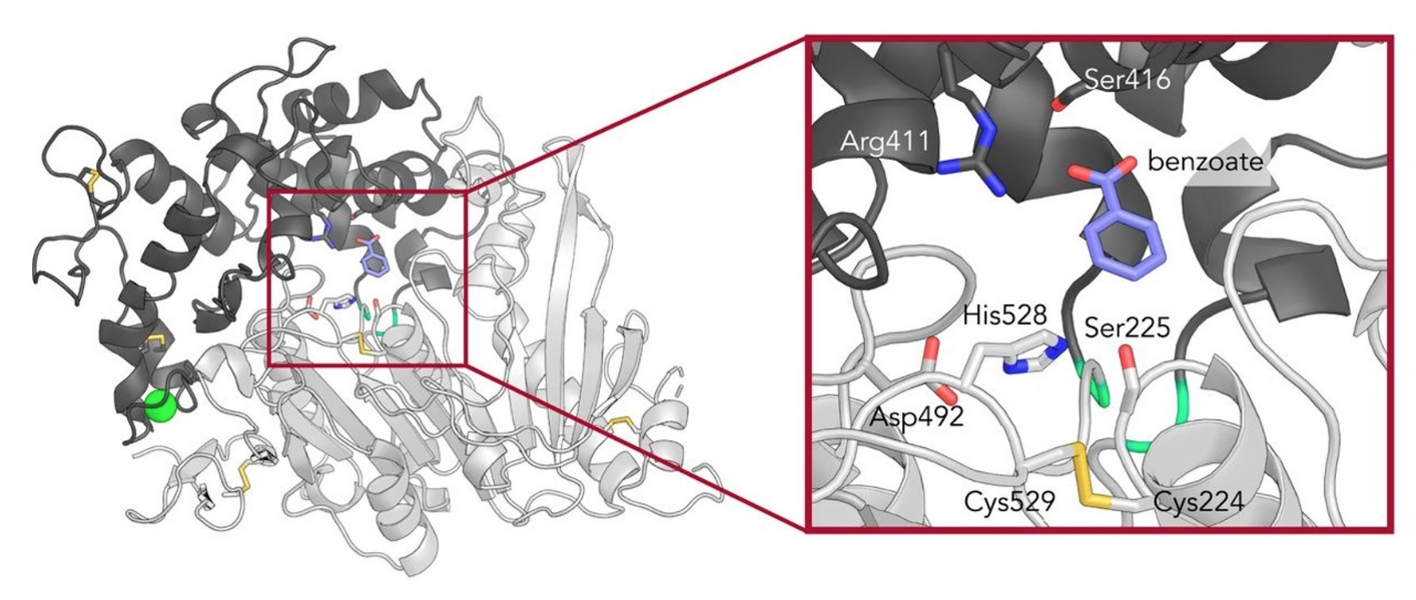
The active site in MHETase works (like some proteases) through a so-called catalytic triad consisting of a histidine, an aspartate and a serine residue. Write a short script that downloads MHETase (PDB-ID: 6QGA) and PETase (PDB-ID: 5XH3) and removes surplus versions of the proteins. Align the two structures with super.
Task 4: Describe the difference between the two structures, find the active site and try to explain why the MHETase has a domain that the PETase does not have, given that the MHETase has a very low kcat.
A hybrid enzyme consisting of an MHETase and a PETase linked to an 8-amino acid residue linker is now prepared. Its ability to form TPA from PET is measured and compared with (1) the PETase alone and (2) the PETase together with the MHETase (in the ratio 1: 1). This gives the following result:

Task 5: Explain the different activity levels of the three enzymes.
Designing enzymes for plastic degradation
Due to the massive problem of plastic pollution worldwide, there is enormous interest in designing new enzymes that degrade plastic. In 2022, a team from Texas published a study about machine learning guided design of a PETase (with accompanying News&Views article).
Task 6: Skim the paper and shortly summarise the rough approach the authors took. Which of the paradigms of machine learning (supervised learning, reinforcement learning, …) did they use? What is the input and output to their model, and what data did they train/test their model on?
Credits
The title image was taken from the PDB-101 resource, where it was presented as the national finalist in the 2019 Wiki Science Competition in the United States.
