Exercise 9b - Nucleic Acids and transcription factors
Exercises | | Links:
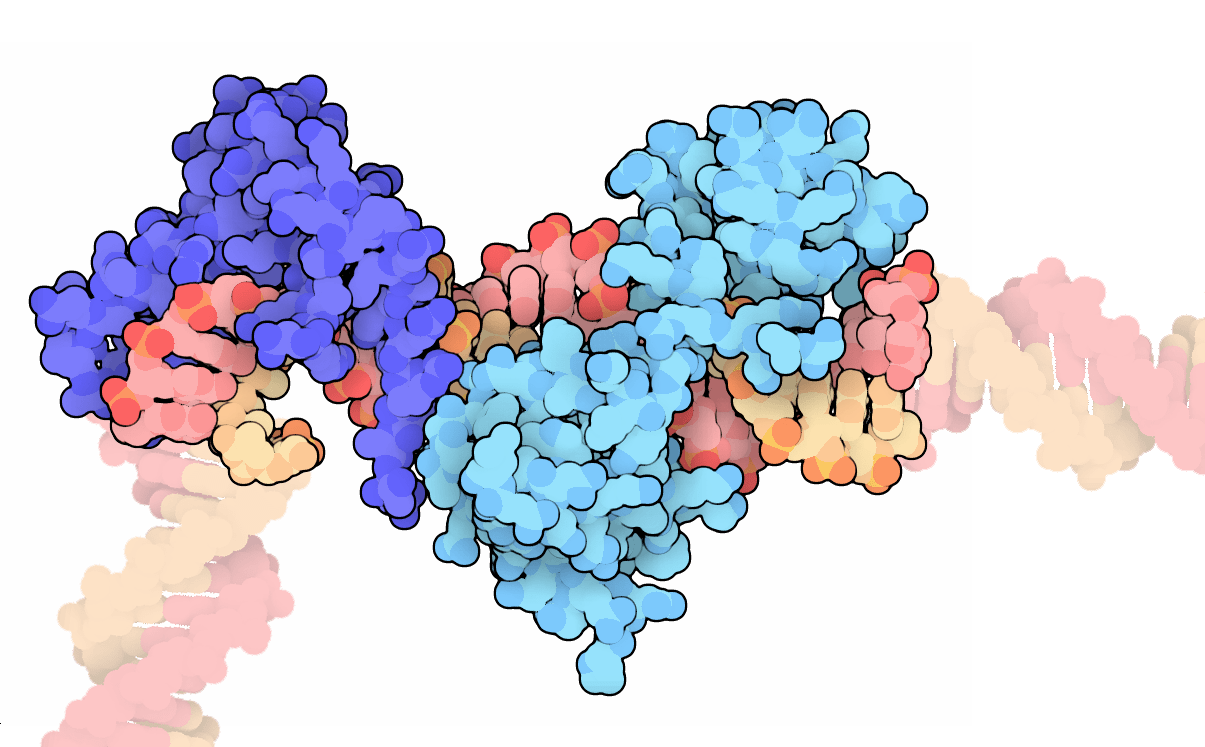
We will learn how DNA and proteins interact with each other.
Introduction
In this assignment you will learn how proteins recognize DNA (or vice versa) in order to enter into a specific (“romantic”) interaction with each other. The secret behind the specific interaction between amino acids and DNA bases is described by the so-called “groove code”. As seen in the figure below, there are exposed H-bond donors and acceptors in both major and minor groove and these are the ones that the amino acids can recognize and bind to.

Task 1: Explain why there is more specificity in binding to major groove than to minor groove. Which amino acids are often used for sequence-specific recognition and why?
The groove code can be a little difficult to spot when analyzing a DNA double helix in PyMOL, but with the right representation it becomes clearer: the DNA-groove.pml script shows the DNA double helix as a spheres where H-bond acceptors are colored red, H-bond donors blue, methyl groups green and non-polar hydrogen atoms yellow.
The sphere representation shows the Van der Waals radius of atoms and thus gives a picture of the realistic shape and volume of the molecule. Coloring of the atoms is seen more clearly due to their size. On the other hand, it is not easy to evaluate the chemical composition (as seen with sticks) or the secondary structure (as seen with cartoon).
Open a DNA double helix with PDB ID: 1BNA in PyMOL and remove water molecules with the PyMOL command remove solvent. Now use the DNA-groove.pml script to change the representation and color the donors and acceptors in the grooves. Rotate the molecule so that you look down into the major groove (so that the symmetry becomes clear).
Task 2: What’s special about the groove code pattern of this double helix DNA?
The TATA-box
The TATA box binding protein (TBP) is involved in promoter recognition to initiate transcription by the RNA polymerase. The TATA box is an 8-base pair region of the promoter that is usually located 30 nucleotides before the transcription start site (TSS). The consensus sequence is TATA(A/T)A(A/T)(G/A). Open the TATA.pml script in PyMOL. F1 shows the binding of TBP along the helical axis of DNA double helix.
Task 3: In which groove of the DNA double helix does TBP bind?
Groove can be identified by zooming in on base pairs and identifying the Högsteen and sugar edge. You can show a base pair using the command “show sticks, resi 7+23” and zoom in to examine it with the commands “zoom resi 7+23” and adjust the depth with the mouse wheel so that the focus is on the base pair. Then rotate so you can see the base pair from the side. Press F2 to display this view.
Use the groove code script (DNA-groove.pml) to display the DNA double helix as a spherical model, where H-bond acceptors are colored red, H-bond donors blue, methyl groups green and non-polar hydrogen atoms yellow. Hide TBP to analyse the groove pattern.
Task 4: What characterizes the groove pattern where TBP binds? What secondary structure element of TBP binds to the DNA groove?
Press F3/F4 to see an important element in the DNA-protein binding, where two phenylalanines intercalate between two base pairs.
Task 5: How are the two base pairs affected by the intercalation of two phenylalanines? Which standard parameter for the relative orientation of bases best describes the conformation of these two base pairs?
Mad-Max
Max-Myc-Mad is a heterodimeric transcription factor system that binds specifically to the DNA segment CACGTG as three different complexes: Myc-Max, Max-Max and Mad-Max. According to the authors of the original article on the Max-Myc-Mad system, the naming of Mad (the second protein in the Mad-Max heterodimer) has nothing to do with the movie of the same name, but comes from the abbreviation of “Max dimerization”. Copy/Paste the PyMOL script Mad-Max.pml and run it in PyMOL. Press F1 to view the Myc-Max complex, press F2 to view the Max-Max complex, and F3 to view the Mad-Max complex.
Task 6: Describe the composition of the dimerization domain and the property of the α-helices that binds DNA. In which groove do Myc-Max, Max-Max and Mad-Max bind?
Orient the molecule in PyMOL so that you look down along the pseudosymmetry axis of Myc-Max and then hide Myc-Max so that only the DNA double helix is visible. Use the groove code script (DNA-groove.pml) to display the groove pattern.
Task 7: What is characteristic of the pattern of H-bond donor and acceptors?
Press F4 to see how the recognition helixes in Myc-Max interact with the DNA segment. Press F5 to see how the recognition helixes in Max-Max interact with the DNA segment.
Press F6 to see how the recognition helix in Mad-Max interacts with the DNA segment.
Task 8: Describe the hydrogen bonds of the three amino acids (His, Glu, Arg) in recognition helices in Myc-Max by specifying which bases and which atoms of the bases are involved in binding. Are there other interactions than with the bases? Are the interactions symmetrical in Myc-Max, Max-Max and Mad-Max and why?
Use the menu Mouse > Selection mode > Atoms to better identify the individual atoms that are part of the hydrogen bonds.
A TALE of DNA
Transcription factor-like effector nucleases (TALENs) bind specific DNA sequences using a series of DNA-binding domains called TALE repeats. TALEN proteins can be designed to recognize and cleave specific DNA sequences and can thus be used for genome editing. TALE proteins are discussed in Stryer “Biochemistry” 9th edition, section 5.4, and the structure can be seen in figure 5.38. Open the attached TALE.pml script in PyMOL. Press F1 and F2 to see the DNA-protein complex from the side and along the helix axis, respectively. Press F3 to see sphere representation, where you can more easily see the repeat structure. Press F4 to see a specific interaction between protein and DNA.
Task 9: Describe the hydrogen bonds of the three amino acids (His, Glu, Arg) in recognition helices in Myc-Max by specifying which bases and which atoms of the bases are involved in binding. Are there other interactions than with the bases? Are the interactions symmetrical in Myc-Max, Max-Max and Mad-Max and why?
Each TALE repeat contains 34 amino acids and two alpha helices, but only two amino acid residues are responsible for the unique recognition of a single nucleotide in the DNA double helix. Press F4-F7 to see how TALE repeats recognize specific nucleotides.
Task 10: Describe which amino acids interact with which bases and what type of interaction it is. Which edge of the bases is involved in the interaction and which of the DNA strands is recognized?
One can design TALE proteins by organizing the order of TALE repeats according to the DNA sequence to be recognized.
Task 11: 3. Suggest which protein functionalities can be put at the end of a TALE protein domain and describe what biological effects it can have and/or what technological possibilities it can provide.
Arc repressor
Here you will analyze DNA-protein binding in PyMOL. You will learn how to color proteins according to secondary structure and how to find polar bonds between two selections.
Arc repressor (PDB ID: 1BDT) is a transcription factor from bacteriophage P22 that inhibits its own gene arc. Arc repressor binds cooperatively as two dimers to a 21-base pair operator site. The DNA operator sequence is shown below, where the TAGA sequence is colored gray. The pseudo-symmetry axis of the inverted repeat passes through the central G-C base pair.
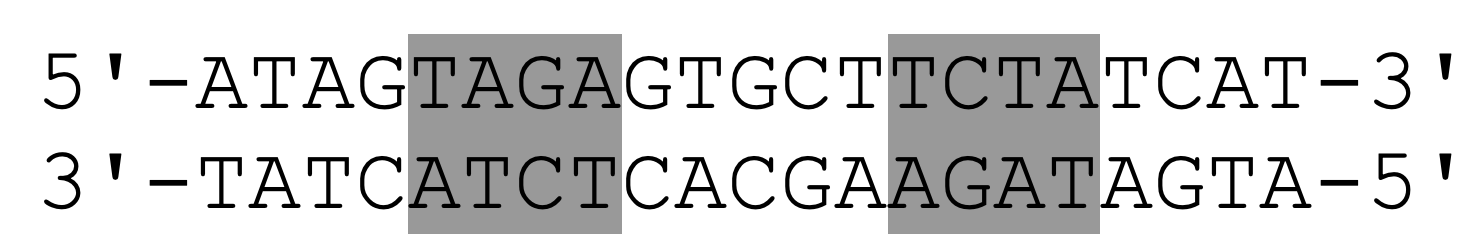
Task 12: Create a script that retrieves the Arc repressor and shows the DNA-protein complex down along the axis of symmetry of the inverted repeat. Use the groove code script (DNA-groove.pml) to format the DNA helix. Save the scene named F1. What is characteristic of the pattern of H-bond donors and acceptors at the two binding sites on DNA and what is this type of recognition sequence called? What is the benefit of this type of binding?
When coloring proteins by secondary structure, there is no single command that does. You have to color each type, alpha helices, beta strands and loops, individually. Loops contain all secondary structures that are not beta strands or alpha helices. You have previously learned that the property selector for secondary structure is ss. PyMOL’s name for alpha helices, beta stands and loops is h, s and l+"", respectively. As an example, you can color loops white with the command color white, ss l+"".
Task 13: Create a new scene named F2 that shows the interaction of DNA with the protein. Describe the binding of the Arc protein to DNA. What type of protein-secondary structure binds to which groove on the DNA double helix? Describe how you observe which groove is being bound.
Interactions between DNA and proteins are best represented if DNA is shown with sticks and colored by element and protein is shown with cartoon and colored by secondary structure.
The distance function can take an extra argument called mode, which has several settings, but the most important are mode = 0 and mode = 2. With the mode setting, you can decide what type of interactions you want to include. Mode = 0 is the default and is just the distance between all points in the two selections. Mode = 2 shows only the polar bonds between two selections. Read more here.
Task 14: Make a scene called F4, which shows hydrogen bonds between recognition element (residues 8-14) and DNA. Which amino acids make H-bonds to the bases? What other types of interactions contribute to binding specificity?
Credits
The title image was taken from the PDB 101 resource, where it was posted to highlight the transcription factors Oct and Sox as molecules of the month.
