Lesson 1 - Introduction
How to use PyMol to dive into the world of proteins
Required reading for this lesson Permalink
Preface to Introduction to Protein Structural Bioinformatics Introduction to Protein Structure
Optional reading for this lesson Permalink
Data Resources for Structural Bioinformatics
Slides Permalink
Video Permalink
This is a course about structural bioinformatics. We will talk about all different kinds of structures, focusing on proteins but also touching on DNA/RNA and carbohydrates. However, before we dive into the meat of the topic, I want you to get a bit of an intuitive understanding of the molecules we are dealing with; that is why we start with the biomolecular visualisation software PyMol. It will also come in handy later when we want to visualise and test the concepts we learn about.
There are different ways to interact with PyMol; one of the most natural ones is the Graphical User Interface (GUI), so basically what you are used to from Word PowerPoint etc: you click buttons and then something happens.
This is different from the command line interface (CLI) that we will use later on. In the CLI, you type commands and then something happens. The CLI is very powerful, but it is also very unforgiving. If you make a typo, you will get an error message and the program will stop. In the GUI, you can click buttons and if you make a mistake, you can just click the undo button and start over.
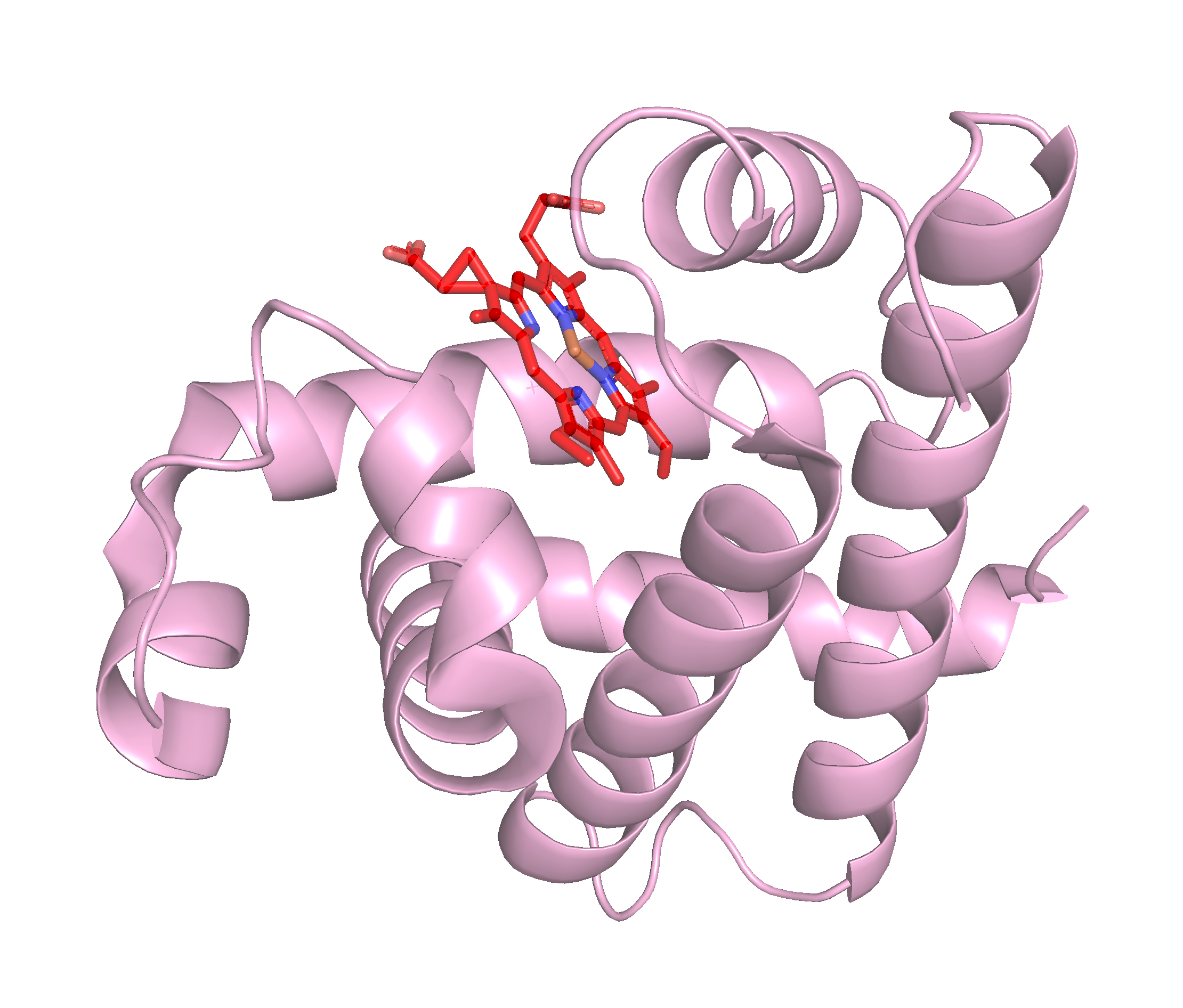
For this lesson, we will start using the GUI to get a feel for PyMol. We will also use it to get a feel for the structure of proteins. To start, we will look at the structure of myoglobin, which is a protein that is found in muscle cells and is responsible for storing oxygen. You can find the structure of myoglobin in the PDB database by searching for the name of the protein. If you are not familiar with the PDB database, you can read more about it here.
You can read more about myoglobin on the PDB101 website, which is a great resource for learning about proteins and their structures and functions!
To start, we will fetch the structure of myoglobin from the PDB database. To do this, we will use the “Get PDB” button in the “File” menu in the upper right hand corner once you opened PyMol. This will open a window where you can enter the PDB code for myoglobin, which is 1mbn. Once you have entered the code, click the “Download” button and the structure will be downloaded and displayed in PyMol.
With the structure of myoglobin loaded, we can start to explore the structure; follow along with the video below to learn how to use the PyMol GUI to explore the structure of proteins such as myoglobin.
Now that you have seen how to use the PyMol GUI to explore the structure of proteins, you can try it out yourself! Try to play around with the myoglobin structure and see what you can find. You can also try to load other proteins from the PDB database and explore their structures. You can find a list of proteins that are related to myoglobin here.
After you have explored the structure of myoglobin, try to complete exercise 0 to test your knowledge of the PyMol GUI.
